- Cleaved Amplified Polymorphic Sequences (CAPS)
- The Derived Cleaved AmplifiedPolymorphic Sequences...
- Amplified Fragment Length Polymorphism (AFLP)
- Restriction Fragment Length Polymorphism (RFLP)
- Random Amplified Polymorphic DNA (RAPD)
- Real-Time qRT-PCR
- Polymerase Chain Reaction (PCR) Illustrations
- Polymerase Chain Reaction (PCR)
- The Nobel Prize in Chemistry 1993
- What is RT PCR?
- How was PCR (polymerase chain reaction) discovered...
- What is the purpose of doing a PCR (polymerase cha...
- How is PCR (polymerase chain reaction) done?
- What is PCR (polymerase chain reaction)?
- NCBI: PCR
- The Polymerase Chain Reaction
- Principle of the PCR
Sunday, March 11, 2018
Everything about Polymerase Chain Reaction
PCR Methods
- Polymerase Chain Reaction (Molecular Info)
- History of Polymerase chain reaction (PCR)--(1)
- History of Polymerase chain reaction (PCR)--(2)
- Regular PCR Procedure
- Videos and Animations for PCR
- Genotyping by PCR
- PCR Primer Design Tools
- PCR Troubleshooting
- About PCR
- Introduction to PCR
- Polymerase chain reaction--PCR
- PCR Animations
- PCR, RT-PCR, and Real Time PCR Tutorials
- Real Time PCR Protocols
- REAL TIME PCR Papers
- Variants of PCR (5)
- RT PCR Protocols
- Web Source of PCR (3)
- Special PCR Protocols
- Variants of PCR (4)
- Variants of PCR (3)
- Important Publications at Cell
- Important Publications at Nature
- Important Publications at Science
- Variants of PCR (2)
- PCR Manual
- PCR SSCP
- PCR RFLP
- Variants of PCR (1)
- AFLP PCR
- Alu-PCR
- Asymmetric PCR
- Colony PCR Protocols
- Competitive and/or Quantitative RT-PCR
- Degenerate PCR
- In Situ PCR
- Differential Display PCR
- Inverse PCR
- Ligation Mediated Suppression PCR
- Long PCR Protocols
- Methylation Specific PCR
- Multiplex PCR
- Nested PCR
- PCR Elisa
- PCR Brochure and Articles
- RACE PCR
- RAPD PCR
- REP-PCR
- Tail PCR
- Touchdown PCR
- Vectorette PCR
- Web Source of PCR (3)
- Web Source of PCR (2)
- Web Source of PCR (1)
Principles of PCR
PCR
amplifies a specific region of a DNA strand (the DNA target). Most PCR methods
amplify DNA fragments of between 0.1 and 10 kilo
base pairs (kbp), although some techniques allow for amplification of
fragments up to 40 kbp in size. The amount of amplified product is
determined by the available substrates in the reaction, which become limiting
as the reaction progresses.
A
basic PCR set-up requires several components and reagents, including:
- a DNA template that
contains the DNA target region to amplify
- a DNA
polymerase, an enzyme that polymerizes new
DNA strands; heat-resistant Taq
polymerase is especially common, as it is more likely to
remain intact during the high-temperature DNA denaturation process
- two DNA primers that are complementary to
the 3' (three prime) ends
of each of the sense and anti-sense strands of the
DNA target (DNA polymerase can only bind to and elongate from a
double-stranded region of DNA; without primers there is no double-stranded
initiation site at which the polymerase can bind); specific primers
that are complementary to the DNA target region are selected beforehand,
and are often custom-made in a laboratory or purchased from commercial
biochemical suppliers
- deoxynucleoside
triphosphates, or dNTPs (sometimes called "deoxynucleotide
triphosphates"; nucleotides containing
triphosphate groups), the building blocks from which the DNA polymerase
synthesizes a new DNA strand
- a buffer solution providing a suitable
chemical environment for optimum activity and stability of the DNA
polymerase
- bivalent cations,
typically magnesium (Mg) or manganese (Mn)
ions; Mg2+ is the most common, but Mn2+ can be used for PCR-mediated DNA mutagenesis, as a higher Mn2+ concentration
increases the error rate during DNA synthesis.
- monovalent cations,
typically potassium (K) ions
The
reaction is commonly carried out in a volume of 10–200 μl in
small reaction tubes (0.2–0.5 ml volumes) in a thermal
cycler. The thermal cycler heats and cools the reaction tubes to achieve
the temperatures required at each step of the reaction (see below). Many modern
thermal cyclers make use of the Peltier effect, which permits both heating
and cooling of the block holding the PCR tubes simply by reversing the electric
current. Thin-walled reaction tubes permit favorable thermal conductivity to
allow for rapid thermal equilibration. Most thermal cyclers have heated lids to
prevent condensation at the top of the reaction tube. Older thermal cyclers
lacking a heated lid require a layer of oil on top of the reaction mixture or a
ball of wax inside the tube.
Scientists Can Make Copies of a Gene through PCR
Once scientists have zeroed in on a specific segment of DNA, how do they produce enough copies of that segment for their research? In most cases, the polymerase chain reaction, or PCR, is their method of choice for quickly generating a sufficient amount of identical genetic material for study and analysis.
Prior to the development of PCR in the 1980s, the primary method for producing many copies of a gene was a relatively time-consuming process known as DNA cloning. This technique involved insertion of the gene of interest into living bacterial cells, which in turn replicated the gene along with their own DNA during the division and replication processes.
What is PCR?
The key element of PCR is heat. Throughout the PCR process, DNA is subjected to repeated heating and cooling cycles during which important chemical reactions occur. During these thermal cycles, DNA primers bind to the target DNA sequence, enabling DNA polymerases to assemble copies of the target sequence in large quantities.
PCR makes it possible to produce millions of copies of a DNA sequence in a test tube in just a few hours, even with a very small initial amount of DNA. Since its introduction, PCR has revolutionized molecular biology, and it has become an essential tool for biologists, physicians, and anyone else who works with DNA.
How does PCR work?
Figure 1: The various components required for PCR include a DNA sample, DNA primers, free nucleotides called ddNTPs, and DNA polymerase.
PCR relies on several key chemical components (Figure 1):
- A small amount of DNA that serves as the initial template or target sequence
- A pair of primers designed to bind to each end of the target sequence
- A DNA polymerase
- Four dNTPs (i.e., dATP, dCTP, dGTP, dTTP)
- A few essential ions and salts
The PCR process then uses these ingredients to mimic the natural DNA replication process that occurs in cells. To automate this process, a machine called a thermocyclerjump-starts each stage of the reaction by raising and lowering the temperature of the chemical components at specific times and for a preset number of cycles.
Step 1: Denaturation
Figure 2: When heated, the DNA strands separate.
Step 2: Annealing
Figure 3: When the solution is cooled, the primers anneal.
Step 3: Extension
Figure 4: DNA polymerase attaches to each primer and assembles dNTPs to build a new strand.
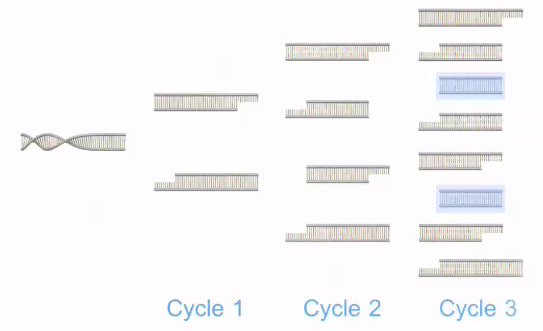
Figure 5: The replication cycle repeats many times.
PCR is an incredibly versatile technique with many practical applications. Once PCR cycling is complete, the copied DNA molecules can be used for cloning, sequencing, mapping mutations, or studying gene expression.
Variations on conventional PCR
Recently, PCR has proven useful in ways beyond merely copying and propagating identical segments of DNA. Today, geneticists rely on PCR to aid in the study of genes themselves.
Copying and quantifying DNA at the same time using real-time PCR
One modification of conventional PCR allows researchers to copy a particular DNA sequence and quantify it simultaneously. Dubbed quantitative real-time PCR (qPCR), this technique makes it possible to measure the amount of DNA produced during each PCR cycle. This refinement involves the use of fluorescent dyes or probes that label double-stranded DNA molecules. These fluorescent markers bind to the new DNA copies as they accumulate, making "real-time" monitoring of DNA production possible.
As the number of gene copies increases with each PCR cycle, the fluorescent signal becomes more intense. Plotting fluorescence against cycle number and comparing the results to a standard curve (produced by real-time PCR of known amounts of DNA) enables scientists to determine the amount of DNA present during each step of the PCR reaction.
Quantifying RNA using reverse transcription PCR
Real-time PCR can also be used to calculate the amount of specific kinds of genetic material other than DNA, such as RNA. This extension of real-time PCR technology, called reverse transcription PCR (RT-PCR), combines real-time PCR with reverse transcription, the process that makes DNA from mRNA. RT-PCR can be used to determine how gene expression changes over time or under different conditions. For this reason, this technique is sometimes used to verify microarray data.
Definition of PCR
Polymerase chain reaction, or PCR, is a laboratory technique used to make multiple copies of a segment of DNA. PCR is very precise and can be used to amplify, or copy, a specific DNA target from a mixture of DNA molecules. First, two short DNA sequences called primers are designed to bind to the start and end of the DNA target. Then, to perform PCR, the DNA template that contains the target is added to a tube that contains primers, free nucleotides, and an enzyme called DNA polymerase, and the mixture is placed in a PCR machine. The PCR machine increases and decreases the temperature of the sample in automatic, programmed steps. Initially, the mixture is heated to denature, or separate, the double-stranded DNA template into single strands. The mixture is then cooled so that the primers anneal, or bind, to the DNA template. At this point, the DNA polymerase begins to synthesize new strands of DNA starting from the primers. Following synthesis and at the end of the first cycle, each double-stranded DNA molecule consists of one new and one old DNA strand. PCR then continues with additional cycles that repeat the aforementioned steps. The newly synthesized DNA segments serve as templates in later cycles, which allow the DNA target to be exponentially amplified millions of times.
https://www.nature.com/scitable/definition/polymerase-chain-reaction-pcr-110
Learn PCR
PCR (short for Polymerase Chain Reaction) is a relatively simple and inexpensive tool that you can use to focus in on a segment of DNA and copy it billions of times over. PCR is used every day to diagnose diseases, identify bacteria and viruses, match criminals to crime scenes, and in many other ways. Step up to the virtual lab bench and see how it works!
Primers are short pieces of DNA that are made in a laboratory. Since they're custom built, primers can have any sequence of nucleotides you'd like.
In a PCR experiment, two primers are designed to match to the segment of DNA you want to copy. Through complementary base pairing, one primer attaches to the top strand at one end of your segment of interest, and the other primer attaches to the bottom strand at the other end. In most cases, 2 primers that are 20 or so nucleotides long will target just one place in the entire genome.
Primers are also necessary because DNA polymerase can't attach at just any old place and start copying away. It can only add onto an existing piece of DNA.
DNA Polymerase is a naturally occurring complex of proteins whose function is to copy a cell's DNA before it divides in two. When a DNA polymerase molecule bumps into a primer that's base-paired with a longer piece of DNA, it attaches itself near the end of the primer and starts adding nucleotides. (In nature, these primers are made by an enzyme called primase).
The DNA polymerase in our bodies breaks down at temperatures well below 95 °C (203 °F), the temperature necessary to separate two complementary strands of DNA in a test tube. The DNA polymerase that's most often used in PCR comes from a strain of bacteria calledThermus aquaticus that live in the hot springs of Yellowstone National Park. It can survive near boiling temperatures and works quite well at 72 °C (162 °F).
Nucleotides are the building blocks that DNA molecules are made of. You add a mixture of four types of nucleotides to your PCR reaction—A's, C's, G's and T's. DNA polymerase grabs nucleotides that are floating in the liquid around it and attaches them to the end of a primer.
JOVE Vieo: PCR: The Polymerase Chain Reaction
The polymerase chain reaction, or PCR, is a technique used to amplify DNA through thermocycling – cyles of temperature changes at fixed time intervals. Using a thermostable DNA polymerase, PCR can create numerous copies of DNA from DNA building blocks called dinucleoside triphosphates or dNTPs. There are three steps in PCR: denaturation, annealing, and elongation. Denaturation is the first step in the cycle and causes the DNA to melt by disrupting hydrogen bonds between the bases resulting in single-stranded DNA. Annealing lowers the temperature enough to allow the binding of oligonucleotide primers to the DNA template. During the elongation step DNA polymerase will synthesize new double-stranded DNA.
This video provides an introduction to the PCR procedure. The basic principles of PCR are described as well as a step-by-step procedure for setting up a generalized PCR reaction. The video shows the necessary components for a PCR reaction, includes instruction for primer design, and provides helpful hints for ensuring successful PCR reactions.
NIH: Introduction of PCR
PCR is a revolutionary method developed by Kary Mullis in the 1980s. PCR is based on using the ability of DNA polymerase to synthesize new strand of DNA complementary to the offered template strand. Because DNA polymerase can add a nucleotide only onto a preexisting 3'-OH group, it needs a primer to which it can add the first nucleotide. This requirement makes it possible to delineate a specific region of template sequence that the researcher wants to amplify. At the end of the PCR reaction, the specific sequence will be accumulated in billions of copies (amplicons).
How It Works
 | Components of PCRDNA template- the sample DNA that contains the target sequence. At the beginning of the reaction, high temperature is applied to the original double-stranded DNA molecule to separate the strands from each other.DNA polymerase- a type of enzyme that synthesizes new strands of DNA complementary to the target sequence. The first and most commonly used of these enzymes isTaqDNA polymerase (fromThermis aquaticus), whereasPfuDNA polymerase (fromPyrococcus furiosus) is used widely because of its higher fidelity when copying DNA. Although these enzymes are subtly different, they both have two capabilities that make them suitable for PCR: 1) they can generate new strands of DNA using a DNA template and primers, and 2) they are heat resistant.Primers- short pieces of single-stranded DNA that are complementary to the target sequence. The polymerase begins synthesizing new DNA from the end of the primer.Nucleotides (dNTPs or deoxynucleotide triphosphates)- single units of the bases A, T, G, and C, which are essentially "building blocks" for new DNA strands.RT-PCR(Reverse Transcription PCR) is PCR preceded with conversion of sample RNA into cDNA with enzymereverse transcriptase.Limitations of PCR and RT-PCR
The PCR reaction starts to generate copies of the target sequence exponentially. Only during the exponential phase of the PCR reaction is it possible to extrapolate back to determine the starting quantity of the target sequence contained in the sample. Because of inhibitors of the polymerase reaction found in the sample, reagent limitation, accumulation of pyrophosphate molecules, and self-annealing of the accumulating product, the PCR reaction eventually ceases to amplify target sequence at an exponential rate and a "plateau effect" occurs, making the end point quantification of PCR products unreliable. This is the attribute of PCR that makesReal-Time Quantitative RT-PCRso necessary.
|
Subscribe to:
Posts (Atom)